|
Seohui Bae I'm a research scientist at LG AI Research in South Korea. At LG AI Research, I've worked on reasoning, out-of-distribution extrapolation, and neural functionals. I completed my bachelor's and master's studies at KAIST, where I was fortunate to be advised by Prof. Eunho Yang. |
|
News
|
ResearchI work at the intersection of reasoning, adaptation, and learning under distribution shifts. My research interests include the following topics:
I regularly contribute to academic publications and collaborative research projects. I’m especially interested in bridging industrial challenges with generalizable solutions in: inference-time scaling, long-tail generalization, and extrapolation |
Interests
|
Education
|
Selected Publications(* equal contribution; † co-corresponding) |
|
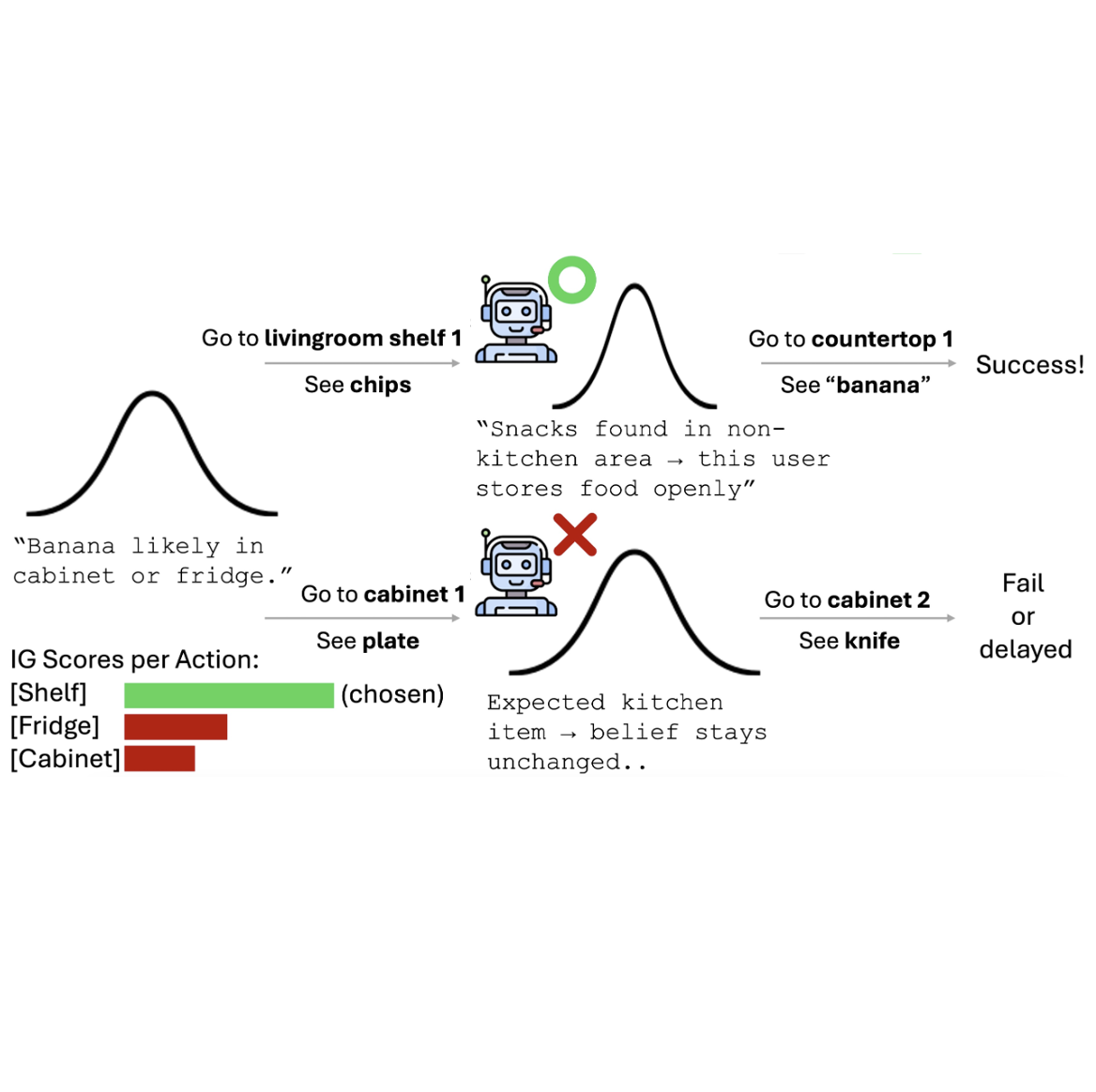
Align While Search: Belief-Guided Exploratory Inference for World-Grounded Embodied Agents
Seohui Bae, Jeonghye Kim, Youngchul Sung, Woohyung Lim preprint, 2025 [pdf] ICML Workshop on Exploration in AI Today, 2025 [pdf] keyword: epistemic exploration, language model agent, test-time adaptation |
|
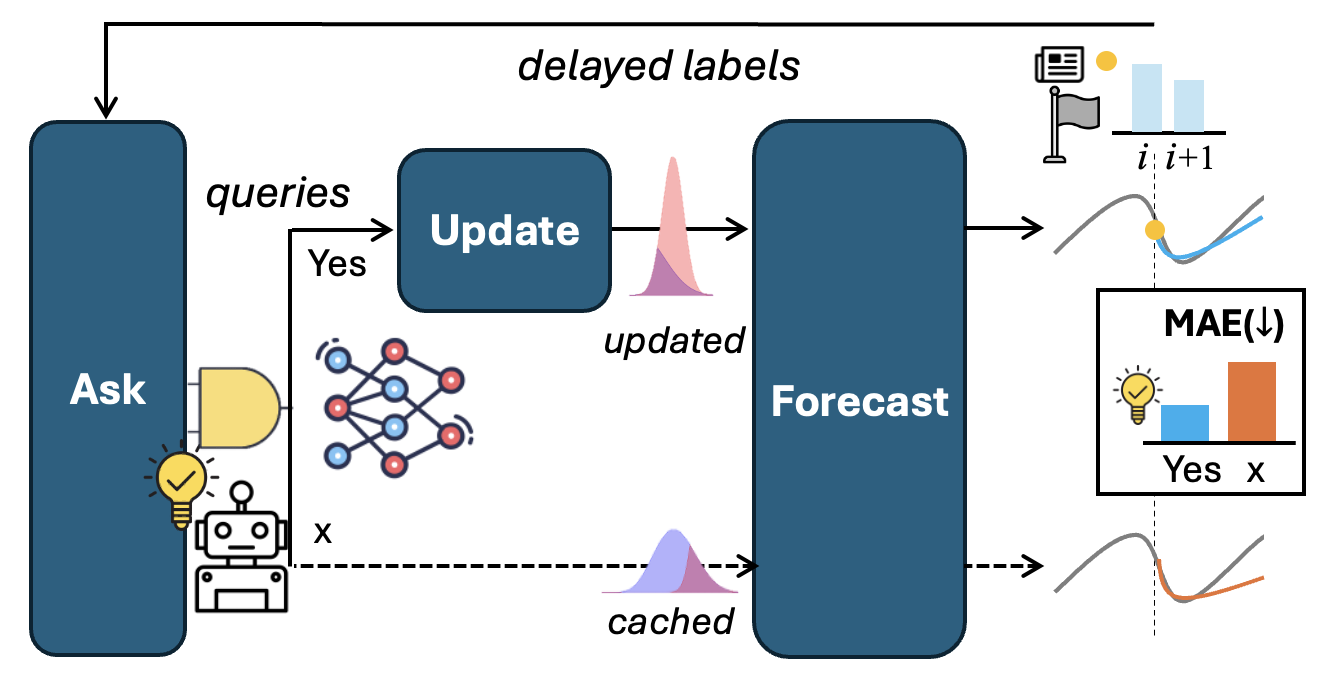
Language-Agent Forecasting with World-Model Surrogates under Delayed Feedback
Seohui Bae, Sangjun Han, Junhyeok Kang, Soyeon Park, Hyeokjun Choe, Soonyoung Lee preprint, 2025 [pdf] keyword: forecasting, language agent, world-model surrogate |
|
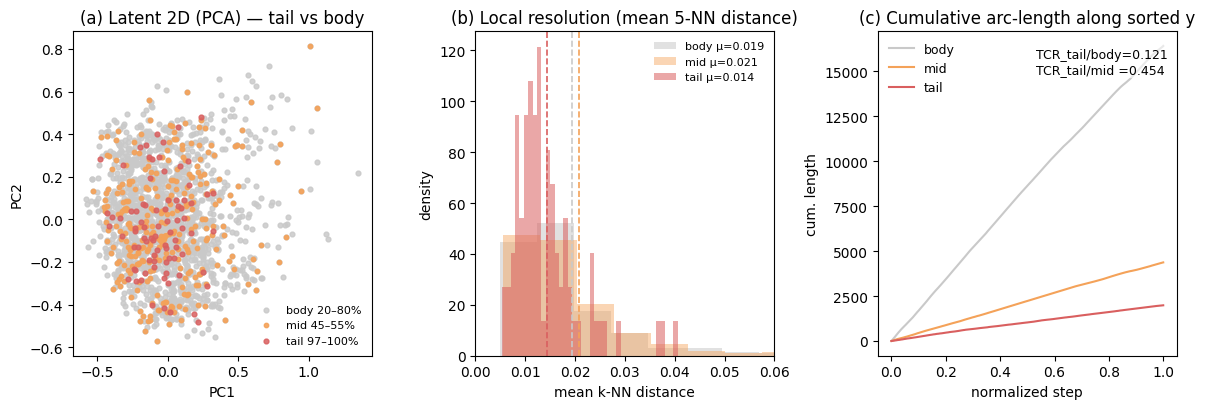
Geometry-Aware Normalization for Imbalanced Time-series Forecasting
Seohui Bae, Junhyeok Kang, Jun Seo, Soyeon Park, Wonbin Ahn, Soonyoung Lee preprint, 2025 [pdf] keyword: time-series, heavy-tail distribution, normalization |
ProjectsLG AI Research
Ongoing Research
|
EducationM.S. in Graduate School of Artificial Intelligence, Mar 2020–Feb 2022
B.S. in Biological Science, Computer Science (minor), Mar 2015–Feb 2020
Korea Science Academy of KAIST, Mar 2012–Feb 2015 |
Academic ServiceConference / Journal Reviewer
|
|
Last date of update: 2025-10-05 / template |